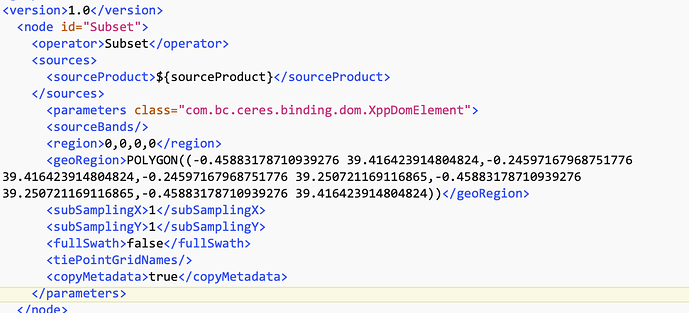
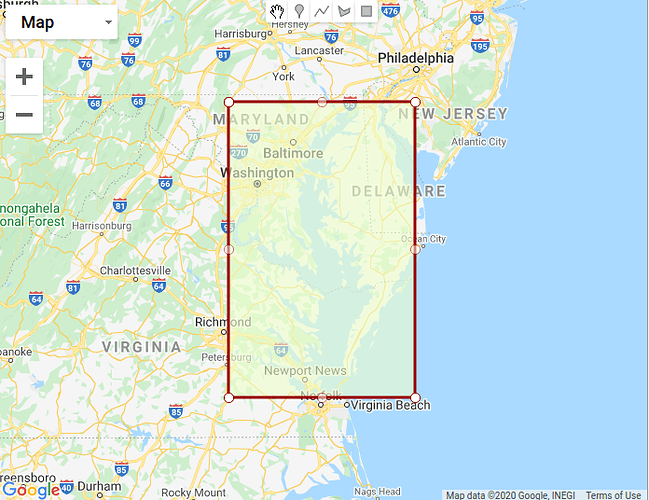
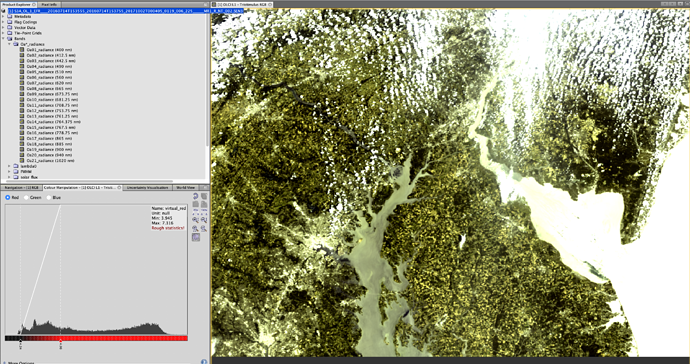
I am trying to use an .xml to batch process 1200 OLCI files. I’d like to subset, apply idepix, and then process using c2rcc. I have tried the below .xml file on 10 files, but they do not seem to be subsetted. Can you let me know what I am doing wrong?
> > <graph id="Graph"> > <version>1.0</version> > > <!-- Subset the product by geographic polygon. --> > <node id="Subset"> > <operator>Subset</operator> > <sources> > <source>${source}</source> > </sources> > <parameters class="com.bc.ceres.binding.dom.XppDomElement"> > <sourceBands/> > <geoRegion>POLYGON ((-77.37755584716797 39.660213470458984, -75.18029022216797 39.660213470458984, -75.18029022216797 36.92744827270508, -77.37755584716797 36.92744827270508, -77.37755584716797 39.660213470458984, -77.37755584716797 39.660213470458984))</geoRegion> > <subSamplingX>1</subSamplingX> > <subSamplingY>1</subSamplingY> > <fullSwath>false</fullSwath> > <tiePointGridNames/> > <copyMetadata>true</copyMetadata> > </parameters> > </node> > > <!-- Calculate IdePix flags across subset scene; these are retained by NOT applied. --> > <node id="Idepix.Olci"> > <operator>Idepix.Olci</operator> > <sources> > <sourceProduct refid="Subset"/> > </sources> > <parameters class="com.bc.ceres.binding.dom.XppDomElement"> > <radianceBandsToCopy/> > <reflBandsToCopy/> > <outputSchillerNNValue>false</outputSchillerNNValue> > <computeCloudBuffer>true</computeCloudBuffer> > <cloudBufferWidth>2</cloudBufferWidth> > <useSrtmLandWaterMask>true</useSrtmLandWaterMask> > </parameters> > </node> > > <!-- Merge IdePix flags back into subset product. --> > <node id="BandMerge"> > <operator>BandMerge</operator> > <sources> > <sourceProduct refid="Subset"/> > <sourceProduct.1 refid="Idepix.Olci"/> > </sources> > <parameters class="com.bc.ceres.binding.dom.XppDomElement"> > <sourceBands/> > <geographicError>1.0E-5</geographicError> > </parameters> > </node> > > <!-- Calculate C2RCC across subset scene flags. Ocean and inland water pixels retained. IdePix flags not applied. --> > <node id="c2rcc.olci"> > <operator>c2rcc.olci</operator> > <sources> > <sourceProduct refid="BandMerge"/> > </sources> > <parameters> > <outputAsRrs>true</outputAsRrs> > <validPixelExpression>!quality_flags.invalid AND (!quality_flags.land || quality_flags.fresh_inland_water)</validPixelExpression> > </parameters> > </node> > > </graph>
Also attached is my .bash fileprocessDataset.bash (1.5 KB)