don’t worry about that - every one is here by choice. But you have to specify your questions if you want answers. So the question “what shall I do” is too general, because this is answered in the tutorials and exercises.
Okay Sir I have a problem to work on matlab just to add paths from stamps and the set path you sent ![]() is not working properly. Please let me know it if it solved. thank you in advanced for your encouragement.
is not working properly. Please let me know it if it solved. thank you in advanced for your encouragement.
please add the matlab directory in your stamps installation to the path, as described here: https://www.youtube.com/watch?v=hgkj1ZruSEk (starting at 1:05)
Otherwise Matlab will not know these commands.
Hi! @ABraun
I try to ask you who are always available 
Why when try to ask you who are always available start step 6 do I have n_trial_wraps = NaN?
what kind of error is it? which file can’t read STAMPS? Is this a serious ERROR?
thanks
Hello every one I cant solve this issue would help me please? I add path through matlab application to my directory of INSAR_Master_Date and re run the terminal and it gives me the same thing matlab: command not found due to this problem I cant run the StaMPs on matlab. I would like to thank you very much for your every single support and suggestions.
@volter29: According to this discussion, this variable is not set by the user but automattically defined by SNAP. If the processing does not stop, I would ignore it and check the results.
@abity: This is not related to stamps, please configure your matlab installation properly:
when I processing it happen like this what is the problem I am trying it a lot of times please help me.
please execute stamps(1,1) and so on from the top folder, not inside PATCH_1
Thank you sir it works but unfortunately I am facing other problem when it runs at step 2 Stamp(2,2)
please just read the error message.
Plus, as mentioned above, please note that Matlab is in fact proprietary software. You need to have a license for add-on toolboxes.
Thanks a lot @ABraun for the answer.
It would appear that it does not affect the final result.
Another question.
How can I visually recognize Unwrapping errors?
I know that I have to use ps_plot_(‘usb’), but what exactly should I look for? can you give me an example or recommend a manual where it explains well what an Unwrapping error is?
thanks a lot
this is hard to see for PS because we don’t see the fringes in the interferogram as clearly as in tradtitional DInSAR approaches, because the PS are connected by a network.
Here is an example where the unwrapping produced errors (phase jumps): Basic Subsidence Formation
These are not as clearly visible for PS, but you can plot all interfefrograms and check if there are patterns in single ifgs which are not contained in others. This can also be due to atmospheric disturbance, but still considered a potential error.
@ABraun You are to kind to everybody trying to get started here, a real hero!
I second your point that he best way to start is just to read through all information sources noted in the StaMPS - Detailled instructions thread.
If anyone has questions about my documentation the best way to contact me is via email, I no longer do InSAR things because I finished my thesis but am still happy to help.
Best regards,
Gijs
thank you for your kind message - happy to hear that. I see that the materials you and Matthias provided were really filling a gap, so I’m convinced that they already helped hundreds of people.
Dear,
really I dont’ know where is the fault, I did the change you gave me (A Braun) )but the problem is still remain . yesterday I processed my data in snap one more time i did a new export from snap to stamps but the same problem.
i hope that you can see what i can not see. thank you a lot.
Its sad that my thesis was never published then there was a possible reference for it, now i just see it as help and dont know how people would be able to reference an unpublished thesis.
Happy that it helped so many people!
is it an option to put it on ResearchGate or Zenodo, for example, so it gets an own DOI and is officially citable?
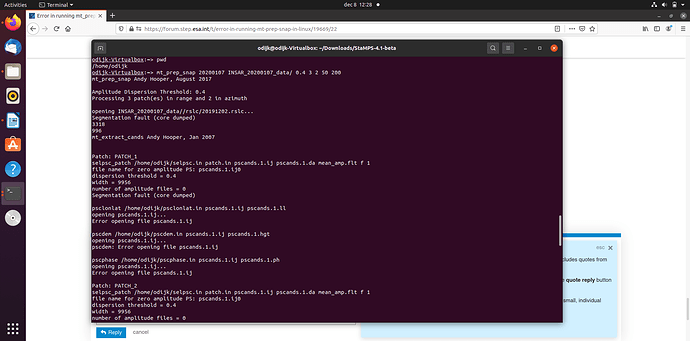
did you see this post? Segmentation Fault (core dumped)